Protein and Ligand Discovery on a Global Scale
The goal of our research program is to advance new platforms for the discovery of chemical probes for human proteins and to use these probes to characterize the functions of proteins in physiology and disease. We have introduced chemical proteomic technologies like activity-based protein profiling (ABPP) to accelerate the discovery of protein functions and small molecules targeting these proteins directly in native biological settings. Much of our current interest involves integrating ABPP methods with emerging knowledge of the genetic basis for human disease to prioritize proteins and pathways for chemical probe development. We are particularly interested in proteins with genetically defined links to cancer, autoimmunity, and nervous system disorders. Our hope is that, by accelerating the discovery of first-in-class chemical probes for human disease-relevant proteins, we will gain mechanistic insights into the functions of these proteins and facilitate the advancement of next-generation therapeutics.
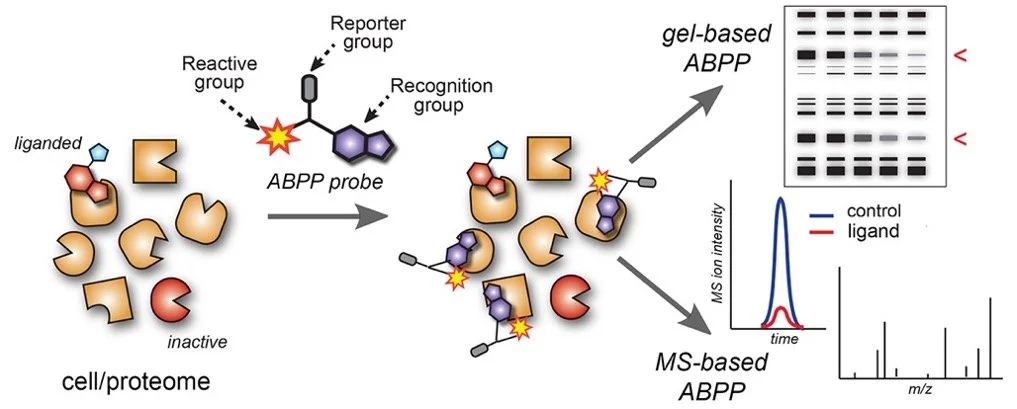
Our lab has pioneered the development of the activity-based protein profiling (ABPP) technology, which uses broad-spectrum chemical probes to functionally characterize large numbers of proteins in native biological systems (Figure 1). Original strategies for ABPP focused on creating chemical probes for specific enzyme families, such as hydrolases, deacetylases, and kinases. These “class-specific” probes are commercially available and remain in widespread use but are restricted to proteins with well-defined active sites. To address this limitation, we have more recently introduced advanced ABPP strategies for simultaneously evaluating the reactivity and small-molecule interactions of thousands of sites on proteins from diverse structural and mechanistic classes.
The activity-based protein profiling (ABPP) technology for protein and ligand discovery. ABPP evaluates the activity and small-molecule interactions of many proteins in parallel in native biological systems using broad-spectrum chemical probes that possess up to three elements: a reactive (electrophilic or photoreactive) and recognition group for targeting functional and druggable sites on proteins, and a reporter group for enriching and identifying probe-modified proteins by gel- and mass spectrometry (MS)-based methods.
Our biological studies with ABPP can be divided into two complementary and cross-fertilizing categories: i) comparative ABPP for target discovery, and ii) competitive ABPP for ligand/probe discovery. In comparative ABPP, multiple biological systems are evaluated to furnish deep maps of differential protein/site (re)activity. Using comparative ABPP, we have functionally annotated orphan, sequence-unrelated members of large enzyme families based solely on their atypical chemical reactivity. In competitive ABPP, the potency and selectivity of candidate small-molecule ligands is assessed across 100s-1000s of proteins in native biological systems. Competitive ABPP has proven particularly useful for covalent ligand and drug discovery, where the technology provides a general platform for optimizing the proteome-wide selectivity of electrophilic compounds. Interestingly, ABPP has also revealed many cryptic sites of functionality/covalent druggability in the human proteome that reside at, for instance, protein-DNA/protein-protein interfaces in transcription factors, scaffolding/adaptor proteins, and E3 ligases. These data thus indicate that combining elements of recognition and reactivity can greatly expand the scope of human proteins targeted by small molecules, especially when screens are performed in native biological settings where the physiological states of proteins are preserved. We collaborate with many synthetic chemistry labs at Scripps and beyond to explore novel reactive groups and chemical scaffolds by ABPP for their interactions with the human proteome.